
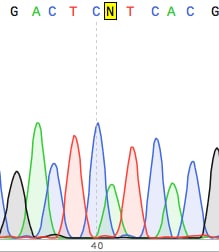
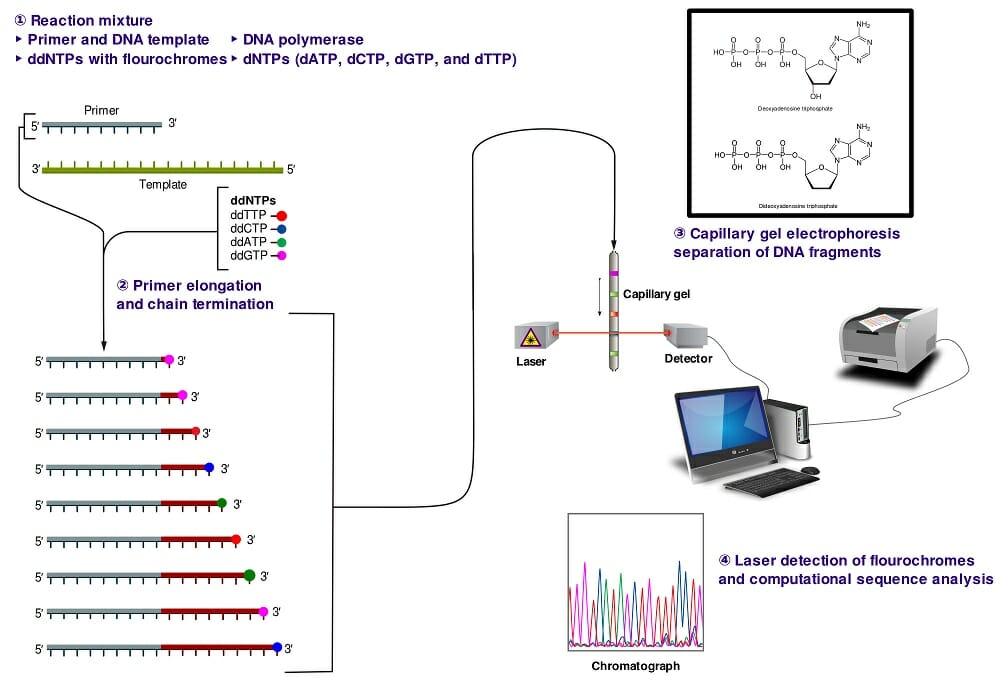
Next-generation sequencing (NGS) is a relatively new method used for sequencing genomes, or portions of genomes, with a high degree of accuracy.
Untangling their profiles and avoiding stutter errors can be exceedingly difficult. All of this changes, however, when samples contain mixtures from more than one person. This is a low number of markers to analyze by modern standards, but it produces a manageable data set that is powerful enough to identify individuals. There is a limit to the number of STRs that can be tested at once, and scientists typically restrict their analyses to about 20 to 30 STRs. Thus, many different STRs are copied and sorted in the same reaction. The more STRs analyzed, the more discriminating the final profile, but doing separate analyses for each STR uses precious amounts of the sample, and can be cost- and time-prohibitive. Indeed, the repetitive nature of STRs can cause a small portion of the PCR product to generate “stutter” - one less or one more of the motif repeats - thus complicating the interpretation of the DNA sample.īesides the introduction of stutter during PCR, traditional STR analysis has other challenges. However, the copying process is known to produce artifacts, making them difficult to read. Making copies of a particular region of the DNA using PCR is one of the most reliable laboratory processes used by genomic scientists. This is easily accomplished in the lab once enough copies of the STRs are available, which is done using polymerase chain reaction (PCR). As different versions of the same STR vary in the number of times its underlying short sequence is repeated, versions can be identified by length. Short Tandem Repeats (STRs) are ideal for human identification, for not only do they vary among individuals more than other genomic regions, but they can be classified without needing to obtain an actual sequence.
#DNA SEQUENCE ANALYSIS SERIES#
Research for the Real World: NIJ Seminar Series.Strategic Challenges and Research Agenda.


 0 kommentar(er)
0 kommentar(er)
